Use the (
) toggle button to choose Y DNA (
) or mitochondrial (mt) DNA (
).
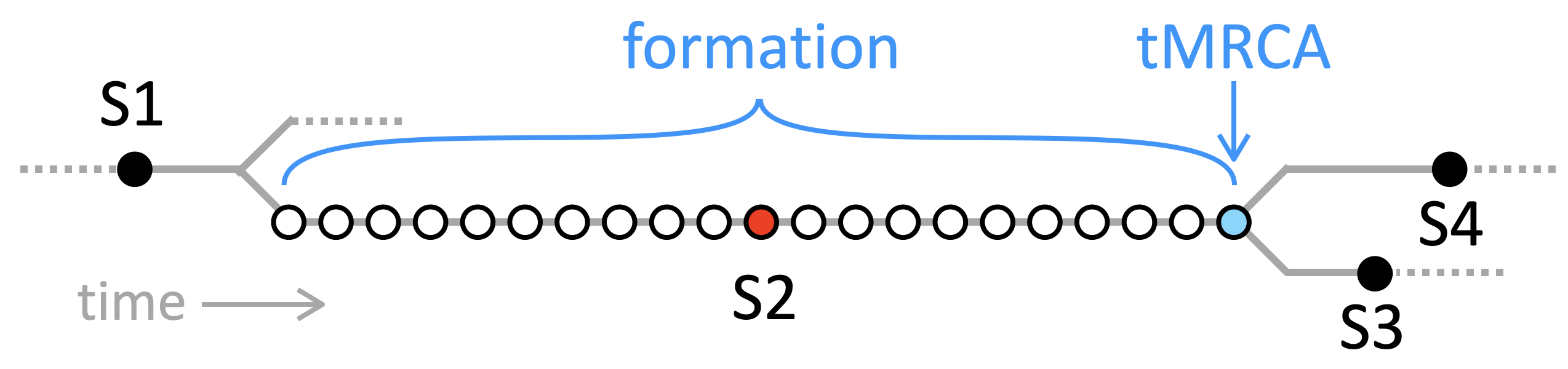
Simply type your haplogroup (ideally your terminal SNP) into the input box and click "Go" or hit return. If this SNP is known to the FTDNA Y or mt Tree, the map will show the path from "Y Adam" or "Mitochondrial Eve" to the SNP. You need type only the lower-level code, for example "M222" instead of the full "R-M222". Y SNP names are as in FTDNA, not the older nomenclature, thus "I-L160" and not "I2a1a1" (although many short 'classic' labels will work, like R1a or G2). If what shows at the top-left of the map is different from what you typed, that's because the tool automatically searches all SNP synonyms and displays the current FTDNA nomenclature. If your haplogroup is missing, then it's not in the FTDNA Y or mt Tree. You may also select examples from the drop-down menu or tap the "shuffle" button () for a random SNP. You can compare paths by entering multiple haplogroups, separated by commas.
Hold down the option/alt key and click () for a random surname (which applies only to Y DNA).